CRAM files are compressed versions of BAM files containing (aligned) sequencing reads. They represent a further file size reduction for this type of data that is generated at ever increasing quantities. Where SAM files are human-readable text files optimized for short read storage, BAM files are their binary equivalent, and CRAM files are a restructured column-oriented binary container format for even more efficient storage.
Tke key components of the approach are that positions are encoded in a relative way (i.e., the difference between successive positions is stored rather than the absolute value) and stored as a Golomb code. Also, only differences to the reference genome are listed instead of the full sequence.
The compression rates achieved are shown in the graph below generated by Uppsala University:
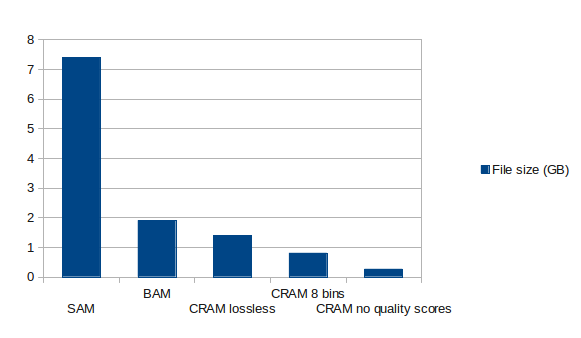
Comparing speed: Using the C implementation of for CRAM (James K. Bonfield), decoding is 1.5–1.7× slower than generating BAM files, but 1.8–2.6× faster at encoding. (File size savings are reported at 34–55%.)
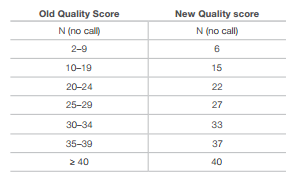
Additional compression can be achieved by reducing the granularity of the quality values which will result in lossy compression though. Illumina suggested a binning of Q scores without significant calling performance.
Binning of similar Q-scores (Illumina):
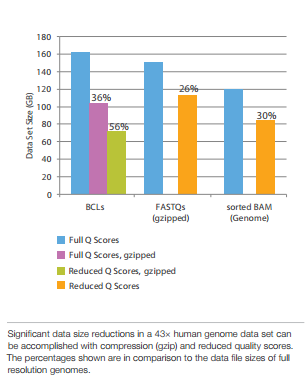
Compression achieved by Q-score binning (Illumina):
Sources and further reading:
- Format definition and usage
- cram-toolkit
- Detailed report at the Uppsala University
- SAMtools with CRAM support
- Original article from Markus Hsi-Yang Fritz, Rasko Leinonen, Guy Cochrane and Ewan Birney
- Article about the implementation in C
- Illumina while paper on Qscore compression
Feature Image: Cram Vectors by Vecteezy


